2026
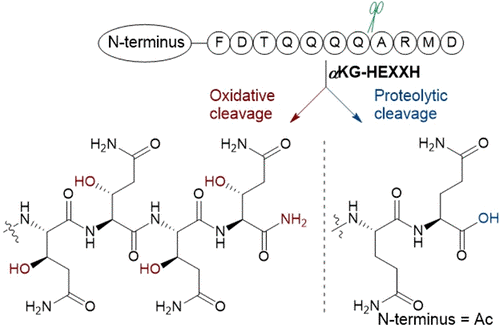
364. Oxidative Peptide Backbone Cleavage by a HEXXH Enzyme During RiPP Biosynthesis
Ouyang, Y.; Yu, Y.; Zhu, L.; Nguyen, D.T.: van der Donk, W.A.* J. Am. Chem. Soc. 2026, ASAP.
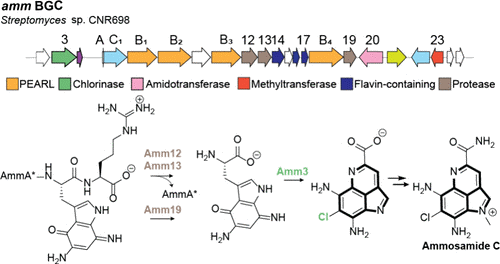
363. The Biosynthetic Pathway to the Pyrroloiminoquinone Marine Natural Product Ammosamide C
Ramos Figueroa, J.; Zhu, L.; Halliman, M.; van der Donk, W.A.* J. Am. Chem. Soc. 2025, 147, 46315-46323.
362. Co-Opting the Bacterial Lipoprotein Pathway in the Biosynthesis of a Lipidated Macrocyclic Peptide
Chen, J.Y.; Zhu, L.; Zhang, K.Y; Berthold, D.A.; van der Donk, W.A.* bioRxiv (2025).
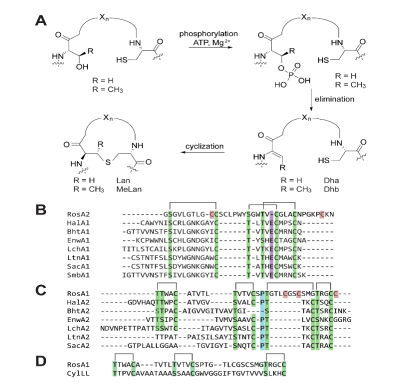
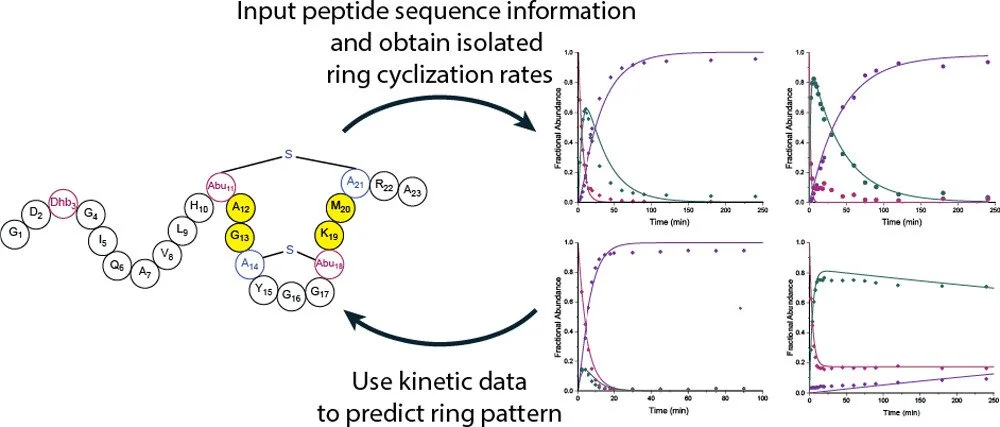
361. An Unusual Ring Pattern in the Rosβ Lanthipeptide of the Two-Component Lantibiotic Roseocin
Desormeaux, E.K.; Zhu, L.; Luo, Y.; Sareen, D.; van der Donk, W.A.*bioRxiv (2025).
2025
360. Aminoacyl-tRNA Specificity of a Ligase Catalyzing Non-Ribosomal Peptide Extension
Nguyen, D.T.; Ramos-Figueroa, J.S.; Vinogradov, A.A.; Goto, Y.; Gadgil, M.G.; Splain, R.A.; Suga, H.; van der Donk, W.A.*; Mitchell, D.A.* (2025). J. Am. Chem. Soc., 147, 37893-37898.
359. Large Protein-Like Leader Peptides Engage Differently with RiPP Halogenases and Lanthionine Synthetases
Vidya, F.N.U.; Luo, Y.; Wu, H.; van der Donk, V.A.; McShan, A.C*; Agarwal, V.* (2025). Nat. Comm., 16, 9273.
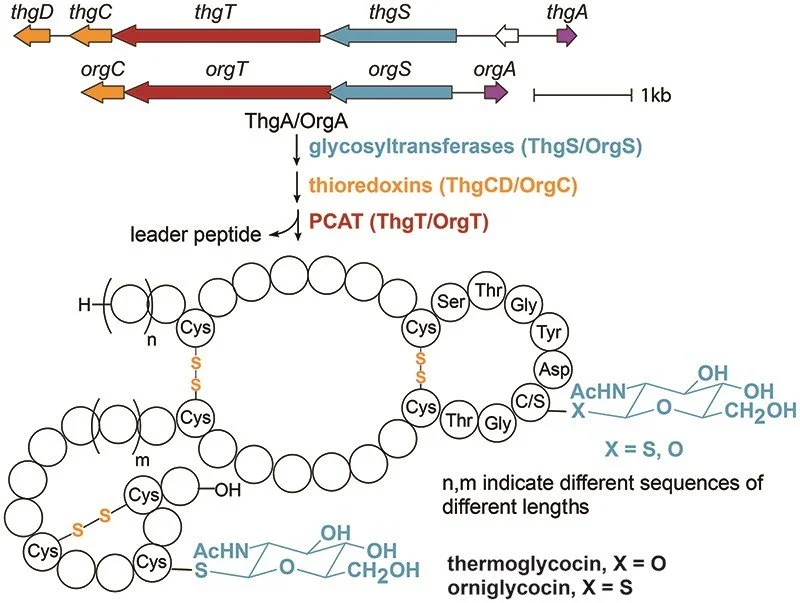
358. Characterization of S-Glycosylated Glycocins Containing Three Disulfides
Martini, R.M.; Padhi, C.; van der Donk, W.A.* (2025). J. Ind. Microbiol. Biotechnol. 52, 1367-5435.
357. Combatting Virulent Gut Bacteria by Inhibiting the Biosynthesis of a Two-component Lanthipeptide Toxin
Moreira, R.; Chakraborty, B.; Yang, Y.; Padhi, C.; 1, Satish K. Nair, S.K.*; van der Donk, W.A.* (2025). Nat. Comm., 16, 6936.
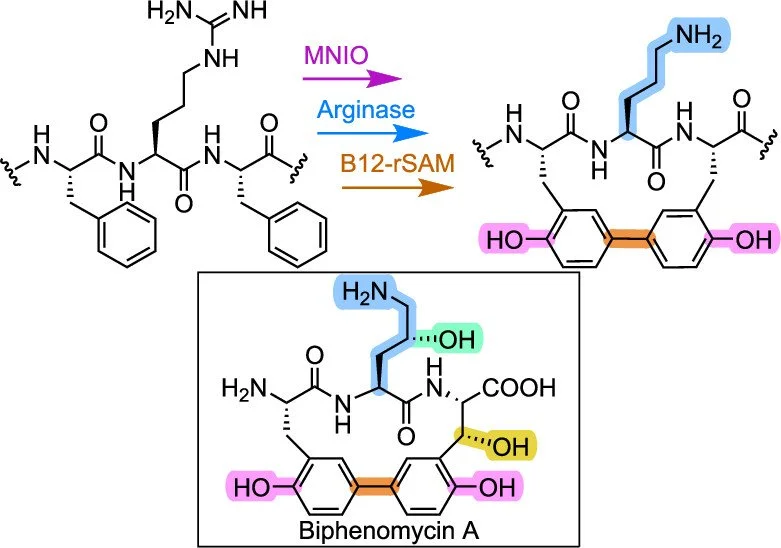
356. Biosynthesis of Biphenomycinlike Macrocyclic Peptides by Formation and Cross-Linking of Ortho-Tyrosines
Padhi, C.; Zhu, L.; Chen J.Y.; Moreira, R.; van der Donk, W.A.* (2025). J. Am. Chem. Soc., 147, 23781-23796.
355. A New Heme Enzyme Family Forms Hydrazine Groups in Diverse Biosynthetic Pathways
Kenney, G.E.; Wang, K-K.A.; Ng, T.L.; van der Donk, W.A.; Balskus, E.P.* (2025). bioRxiv. DOI: 10.1101/2025.05.19.655019.
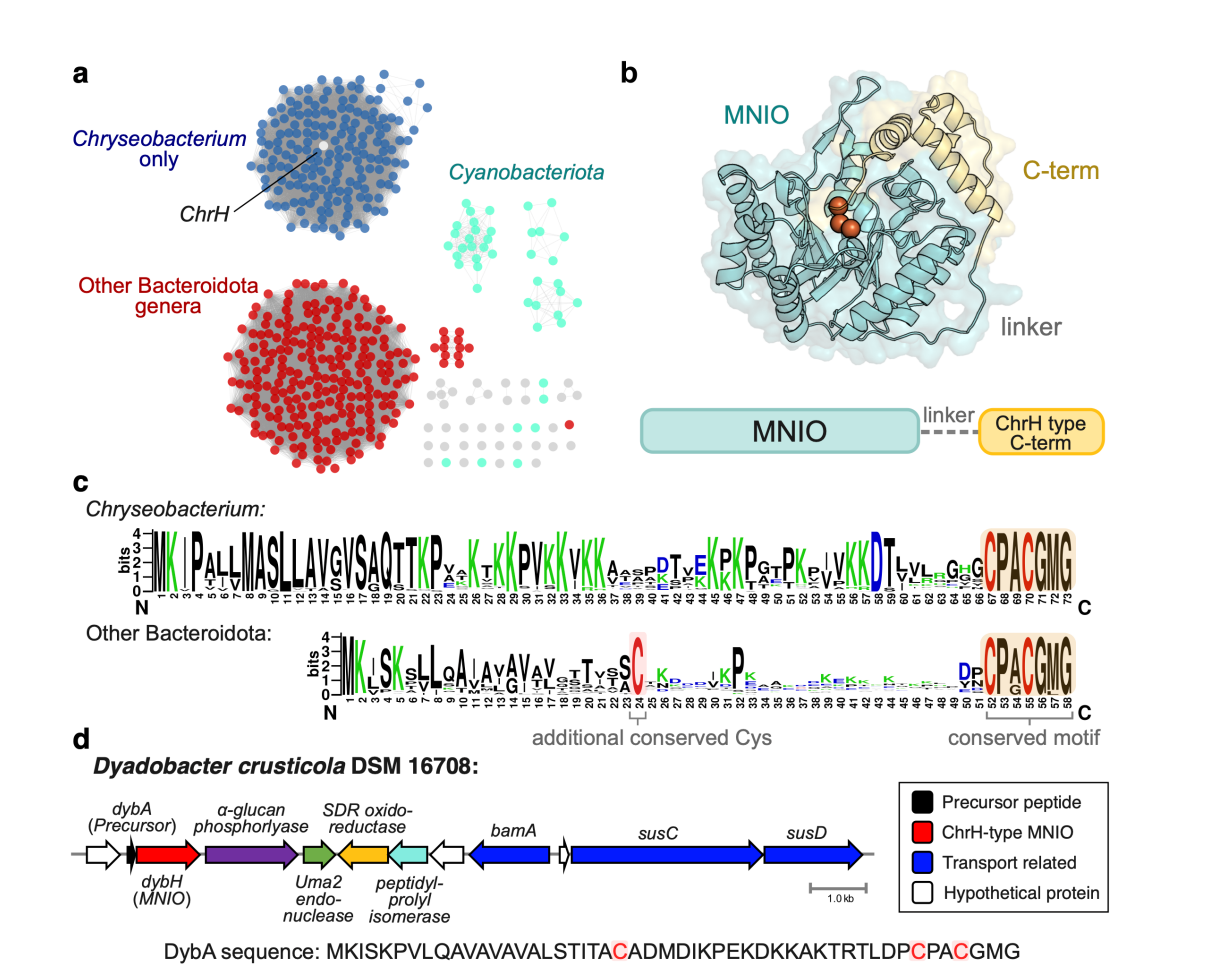
354. Genome Mining for Natural Products Made by Multinuclear Iron-dependent Oxidation Enzymes (MNIOs)
Yu, Y.; van der Donk, W.A.* (2025). Methods in Enzymology, 717, 89-117.
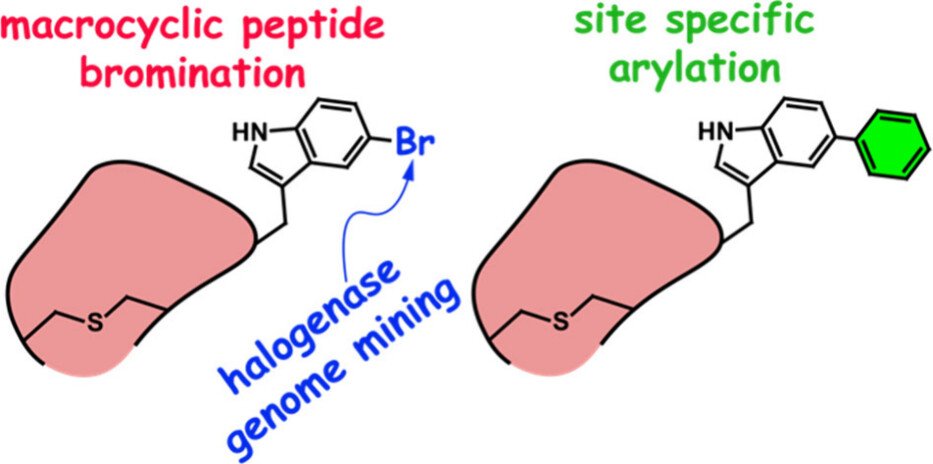
352. Transformation-Guided Genome Mining Provides Access to Brominated Lanthipeptides
Saha, N.; Vidya, F.N.U.; Luo, Y.; van der Donk, W.A.; Agarwal, V.* (2025). Organic Letters, 27, 984-988.
2024
351. Kinetic Analysis of Cyclization by the Substrate-Tolerant Lanthipeptide Synthetase ProcM
Desormeaux, E.K.; Barksdale, G.; van der Donk, W.A.*(2024). ACS Catalysis, 14, 18310-18321.
350. Substrate Specificity of a Methyltransferase Involved in the Biosynthesis of the Lantibiotic Cacaoidin
Liang, H.; Luo, Y.; van der Donk, W.A.* (2024). Biochemistry, 63, 2493-2505.
349. Bibacillin 1: a two-component lantibiotic from Bacillus thuringiensis
Moreira, R.; Yang, Y.; Luo, Y.; Gilmore, M.S.; van der Donk, W.A.* (2024). RSC Chem Biol., 5, 1060-1073.
348. Initial Characterization of the Viridisins’ Biological Properties
Vermeulen, R.*; Du Preez van Staden, A.; Ollewagen, T.; van Zyl, L.J.; Luo, Y.; van der Donk, W.A.; Dicks, L.M.T.; Smith, C.; Trindade, M. (2024). ACS Omega, 9, 31832-31841.
347. Use of a head-to-tail peptide cyclase to prepare hybrid RiPPs
Le, T.; Zhang, D.; Martini, R.M.; Biswas, S.; van der Donk, W.A.*(2024). Chem. Comm., 60, 6508-6511.
346. Multinuclear Non-Heme Iron Dependent Oxidative Enzymes (MNIOs) Involved in Unusual Peptide Modifications
Chen, J.Y.; van der Donk, W.A.* (2024). Curr. Opin. Chem. Biol., 80, 102467.
345. Unexpected Transformations during Pyrroloiminoquinone Biosynthesis
Ramos Figueroa, J.; Zhu, L.; van der Donk, W.A.* (2024). J. Am. Chem. Soc., 146, 14235-14245.
344. Biosynthesis of Macrocyclic Peptides with C-Terminal β-Amino-α-keto Acid Groups by Three Different Metalloenzymes
Nguyen, D.T.; Zhu, L.; Gray, D.L.; Woods, T.J.; Padhi, C.; Flatt, K.M.; Mitchell, D.A.*; van der Donk, W.A.* (2024). ACS Cent. Sci., 10, 1022-1032.
343. Genome Mining for New Enzyme Chemistry
Nguyen, D.; Mitchell, D.A.*; van der Donk, W.A.* (2024). ACS Catal., 14, 4536-4553.
342. Activity of Gut-Derived Nisin-like Lantibiotics against Human Gut Pathogens and Commensals
Zhang, Z.J.*; Wu, C.; Moreira, R.; Dorantes, D.; Pappas, T.; Sundararajan, A.; Lin, H.; Pamer, E.G*; van der Donk, W.A. (2024). ACS Chem. Biol., 19, 357–369.
341. Facile Method for Determining Lanthipeptide Stereochemistry
Luo, Y.; Xu, S.; Frerk, A.; van der Donk, W.A.* (2024). Anal. Chem., 96, 1767–1773.
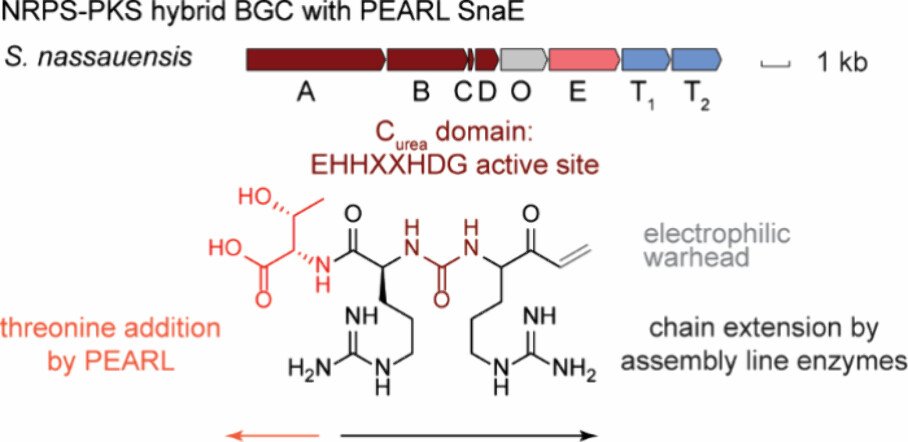
340. PEARL-Catalyzed Peptide Bond Formation after Chain Reversal by Ureido-Forming Condensation Domains
Yu, Y.; van der Donk, W.A.* (2024). ACS Cent. Sci., 10, 1242-1250.
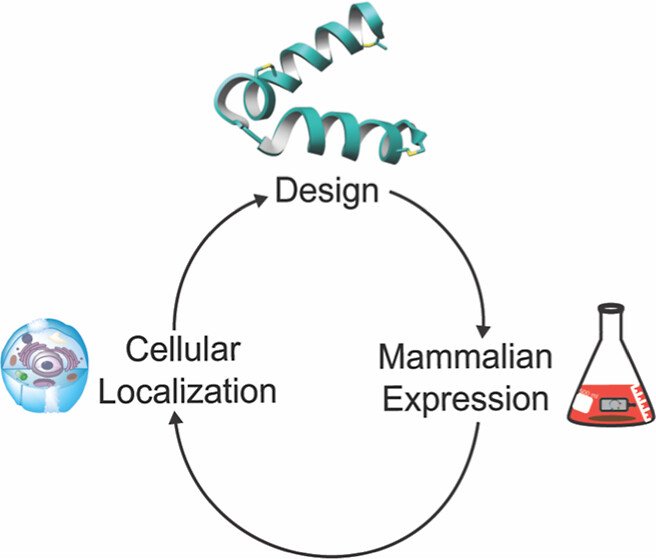
339. Expression and Subcellular Localization of Lanthipeptides in Human Cells
Eslami, S.M.; Padhi, C.; Rahman, I.R.; van der Donk, W.A.* (2024). ACS Synth. Biol., 13, 2128-2140.
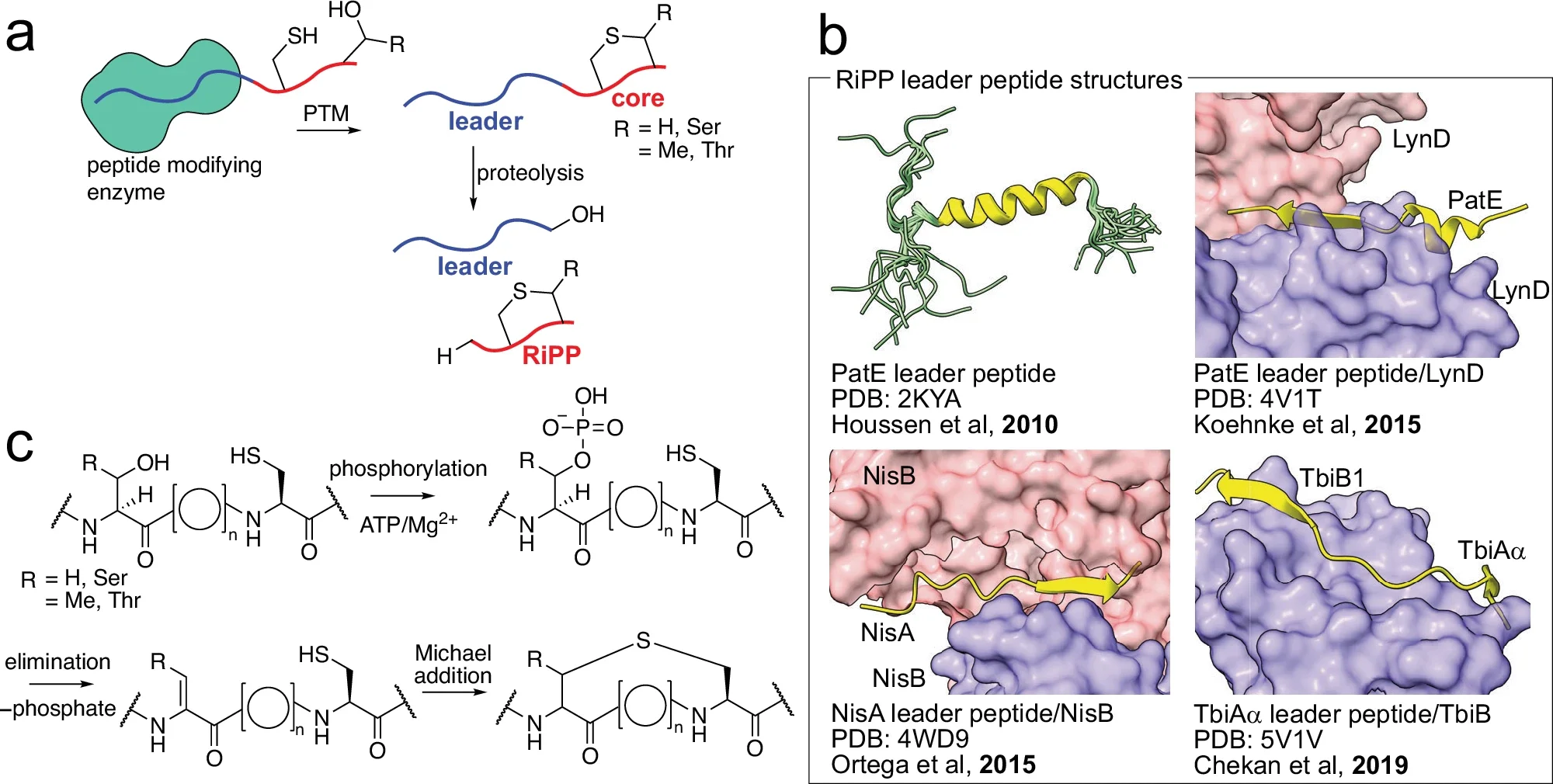
338. Proteases Involved in Leader Peptide Removal during RiPP Biosynthesis
Eslami, S.; van der Donk, W.A.* (2024). ACS Bio Med Chem Au, 4: 20-36.
2023
337. Investigation into the Mechanism of Action of the Antimicrobial Peptide Epilancin 15X
Wu, C.; Lower, B.A.; Moreira, R.; Dorantes, D.; Le, T.; Giurgiu, C.; Shi, Y.; van der Donk, W.A.* (2023). Front. Microbiol., 14: 1247222.
336. Structures of the Holoenzyme TglHI Required for 3-Thiaglutamate Biosynthesis
Zheng, Y.; Xu, X.; Fu, X.; Zhou, X.; Dou, C.; Yu, Y.; Yan, W.; Yang, J.; Xiao, M.; van der Donk, W.A.; Zhu, X.; Cheng, W.* (2023). Structure, 31, 1220–1232.
335. The Circular Bacteriocin Enterocin NKR-5-3B Has an Improved Stability Profile Over Nisin
Wang, C.K.; Huang, Y.-H.; Shabbir, F.; Pham, H.T.; Nicole Lawrence, N.; Benfield, A.H.; van der Donk, W.A.; Henriques, S.T.; Turner, M.S.; David J Craik, D.J.* (2023). Peptides, 167, 171049.
334. Macrocyclization and Backbone Rearrangement During RiPP Biosynthesis by a SAM-Dependent Domain-of-Unknown-Function 692
Ayikpoe, R.S; Zhu, L.; Chen, J.Y.; Ting, C.P.; van der Donk, W.A.* (2023). ACS Cent. Sci., 9, 1008-1018.
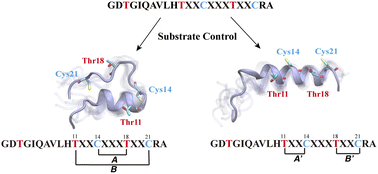
333. Sequence Controlled Secondary Structure Is Important for the Site-selectivity of Lanthipeptide Cyclization
Mi, X.; Desormeaux, E.K.; Le, T.L.; van der Donk, W.A.*; Shukla, D.* (2023). Chem. Sci., 14, 6904-6914.
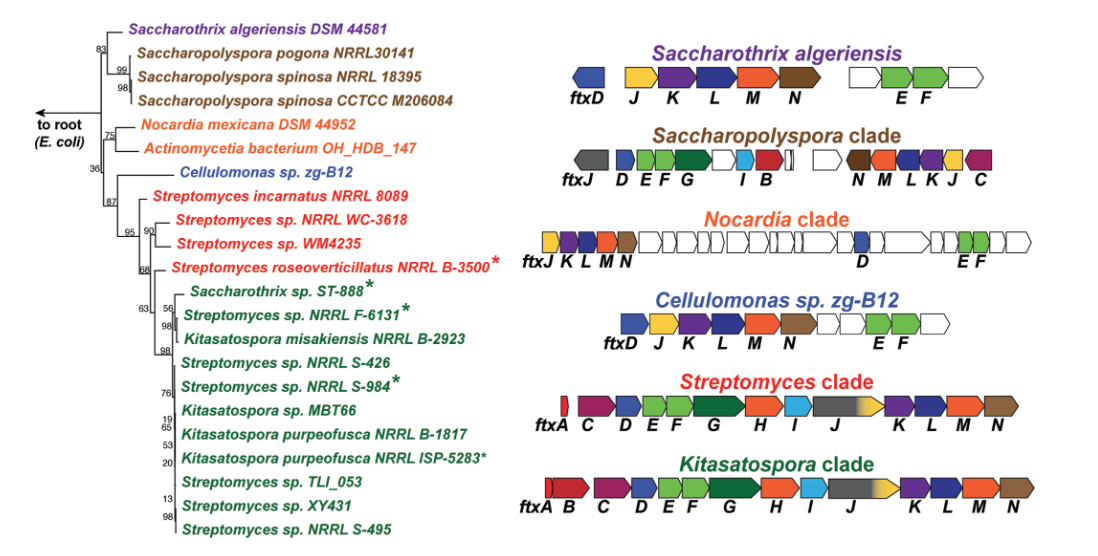
332. A Novel Pathway for Biosynthesis of the Herbicidal Phosphonate Natural Product Phosphonothrixin Is Widespread in Actinobacteria
Bown, L.; Hirota, R.; Goettge, M.; Cui, J.; Krist, D.; Zhu, L.; Giurgiu, C.; van der Donk, W.A.; Ju, K.-S.; Metcalf, W.W.* (2023). J. Bacteriol., 205, e00485-22.
331. Synthesis of Fluorescent Lanthipeptide Cytolysin S Analogs by Late-Stage Sulfamidate Ring-Opening
Mazo, N.; Rahman, I.R.; Navo, C.D.; Peregrina, J.M.; Busto, J.H.; van der Donk, W.A.; Jiménez-Osés, G.* (2023) Org. Lett., 25, 1431-1435.
330. Improved production of class I lanthipeptides in Escherichia coli
Lee, H.; Wu, C.; Desormeaux, E.K.; Sarksian, R.; van der Donk, W.A.* (2023) Chem. Sci., 14, 2537-2546.
329. The Mechanism of thia-Michael Addition Catalyzed by LanC Enzymes
Ongpipattanakul, C.; Liu, S.; Luo, Y.; Nair, S.K.*; van der Donk, W.A.*(2023) Proc. Natl. Acad. Sci. USA, 120, e2217523120.
328. syn-Elimination of Glutamylated Threonine in Lanthipeptide Biosynthesis
Sarksian, R.; Zhu, L.; van der Donk, W.A.* (2023) Chem. Commun., 59, 1165-1168.
2022
327. Class V Lanthipeptide Cyclase Directs the Biosynthesis of a Stapled Peptide Natural Product
Pei, Z.-F.; Zhu, L.; Sarksian, R.; van der Donk, W.A.*; Nair, S.K.* (2022) J. Am. Chem. Soc., 144, 17549-17557.
326. A Scalable Platform to Discover Antimicrobials of Ribosomal Origin
Ayikpoe, R.S; Shi, C.; Battiste, A.J.; Eslami, S.M.; Ramesh, S.; Simon, M.A.; Bothwell, I.R.; Lee, H.; Rice, A.J.; Ren, H.; Tian, Q.; Harris, L.A.; Sarksian, R.; Zhu, L.; Frerk, A.M.: Precord, T.W.; van der Donk, W.A. *; Mitchell, D.A.*; Huimin Zhao, H.* (2022) Nat. Commun., 13, 6135.
325. Mechanism of Action of Ribosomally Synthesized and Post-Translationally Modified Peptides
Ongpipattanakul, C.; Desormeaux, E.K.; DiCaprio, A.; van der Donk, W.A.*; Mitchell, D.A.*; Nair, S.K.* (2022) Chem. Rev., 122, 14722-14814.
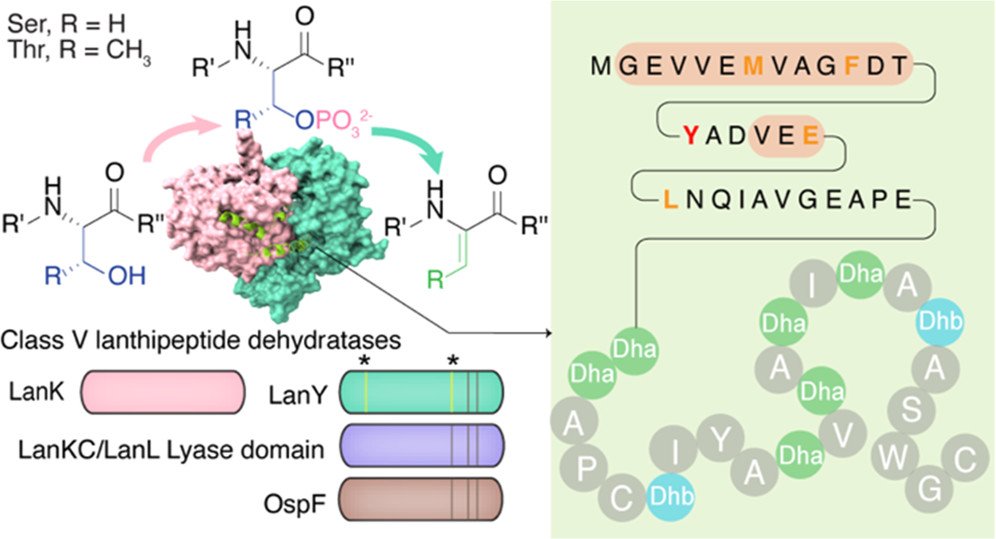
324. Mechanistic Studies on Dehydration in Class V Lanthipeptides
Liang, H.; Lopez, I.J.; Sánchez-Hidalgo, M.; Genilloud, O.; van der Donk, W.A.* (2022) ACS Chem. Biol., 17, 2519-2527.
323. Divergent Evolution of Lanthipeptide Stereochemistry
Sarksian, R.; van der Donk, W.A.* (2022) ACS Chem. Biol., 17, 2551-2558.
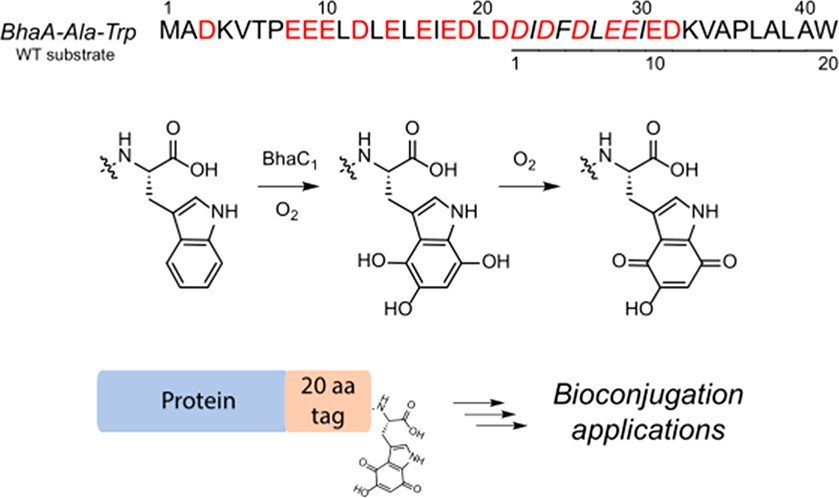
322. Substrate Specificity of the Flavoenzyme BhaC1 that Converts the Indole of Trp into a Hydroxyquinone
Daniels, P.N.; van der Donk, W.A.* (2023) Biochemistry, 62, 378-387.
321. Biosynthesis of 3-Thia-α-Amino Acids on a Carrier Peptide
Yu, Y.; van der Donk, W.A.*(2022) Proc. Natl. Acad. Sci., 29, e2205285119.
320. Accessing Diverse Pyridine-based Macrocyclic Peptides by a Two-Site Recognition Pathway
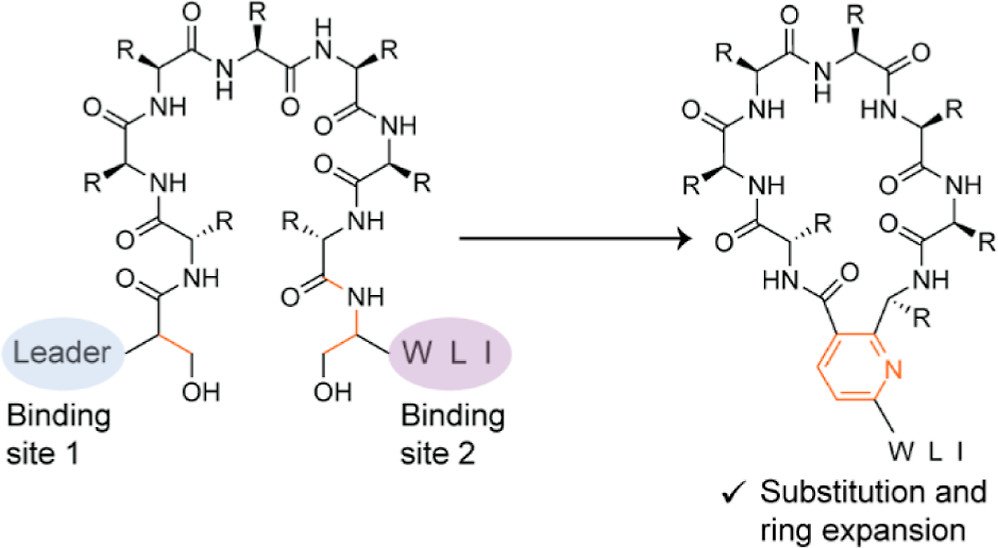
Nguyen, D.T.; Le, T.T.; Rice, A.J.; Hudson, G.A.; van der Donk, W.A.*; Mitchell, D.A.* (2022) J. Am. Chem. Soc., 144, 11263-11269.
319. Macrocyclization and Backbone Modification in RiPP Biosynthesis
Lee, H.; van der Donk, W.A.* (2022) Ann. Rev. Biochem., 91, 269-294.
318. Substrate Recognition by the Peptidyl-(S)-2-Mercaptoglycine Synthase TglHI During 3-Thiaglutamate Biosynthesis
McLaughlin, M.I.; Yu, Y.; van der Donk, W.A.* (2022) ACS Chem. Biol., 17, 930-940.
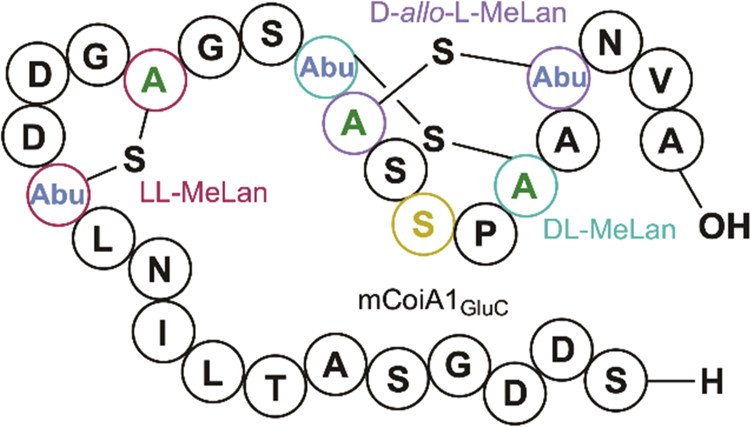
317. Unexpected Methyllanthionine Stereochemistry in the Morphogenetic Lanthipeptide SapT
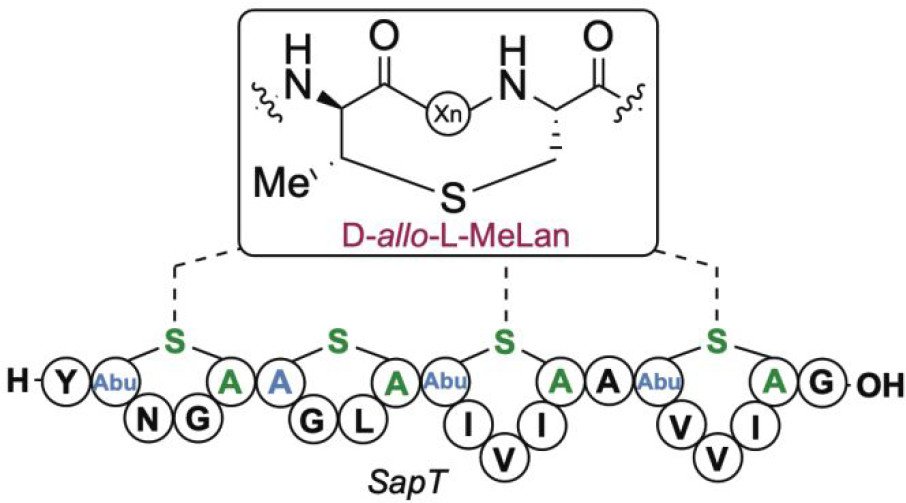
Sarksian, R.; Hegemann, J.D.; Simon, M.A.; Acedo, J.Z.; van der Donk, W.A.* (2022) J. Am. Chem. Soc., 144, 6373-6382.
316. Mechanism of Radical S-Adenosyl-l-methionine Adenosylation: Radical Intermediates and the Catalytic Competence of the 5'-Deoxyadenosyl Radical
Lundahl, M.N.; Sarksian, R.; Yang, H.; Jodts, R.J.; Pagnier, A.; Smith D.F.; Mosquera, M.A.; van der Donk, W.A.; Hoffman, B.M.; Broderick, W.E.; Broderick, J.B.* (2022) Chemical Science, 14(10), 2537–2546.
315. A biosynthetic pathway to aromatic amines that uses glycyl-tRNA as nitrogen donor
Daniels, P.N.; Lee, H.; Splain, R.A.; Ting, C.P.; Zhu, L.; Zhao, X.; Moore, B.S.; van der Donk, W.A.* (2022) Nature Chem., 14, 71-77.
2021
314. Structural and Mechanistic Investigations of Protein S-Glycosyltransferases
Fujinami, D.; Garcia de Gonzalo, C.; Biswas, S.; Hao, Y.; Wang, H.; Garg, N.; Lukk, T.; Nair, S.K.*; van der Donk, W.A.* (2021) Cell Chem. Biol., 28, 1740-1749.
313. Substrate Sequence Controls Regioselectivity of Lanthionine Formation by ProcM
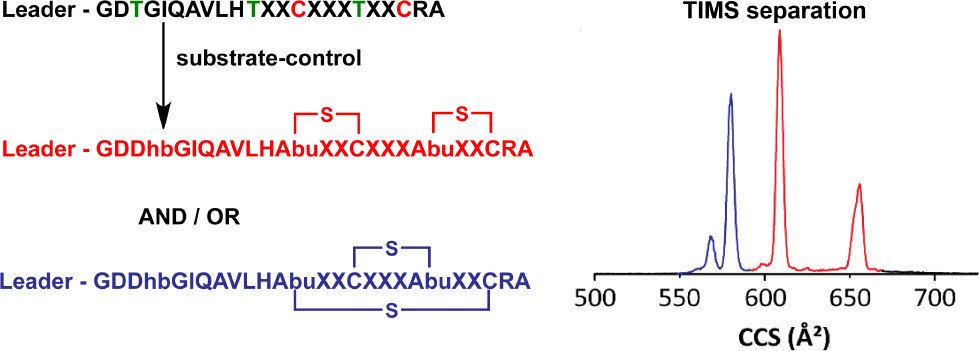
Le, T.; Fouque, K.J.D.; Santos-Fernandez, M.; Navo, C.D.; Jiménez-Osés, G.; Sarksian, R.; Fernandez-Lima, F.*; van der Donk, W.* (2021) J. Am. Chem. Soc., 143, 18733-18743.
312. Structure-Activity Relationships of Enterococcal Cytolysin
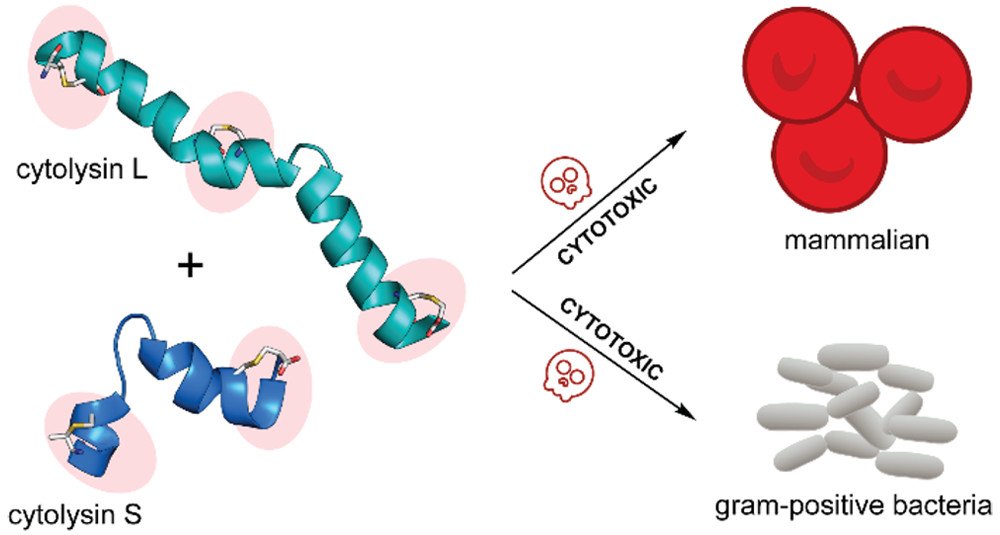
Rahman, I.R.; Sanchez, A.: Tang, W.; van der Donk, W.A.* (2021) ACS Infect. Dis., 7, 2445-2454.
311. The Antimicrobial Activity of the Glycocin Sublancin Is Dependent on an Active Phosphoenolpyruvate-Sugar Phosphotransferase System
Biswas, S.; Wu, C.; van der Donk, W.A.* (2021) ACS Infect. Dis., 7, 2402-2412.
310. Exploring Structural Signatures of the Lanthipeptide Prochlorosin 2.8 Using Tandem Mass Spectrometry and Trapped Ion Mobility-Mass Spectrometry
Jeanne Dit Fouque, K.; Hegemann, J.; Santos-Fernandez, M.; Le, T.; Gomez-Hernandez, M.; van der Donk, W.; Fernandez-Lima, F.* (2021) Anal. Bioanal. Chem., 413, 4815-4824.
309. Peptide Backbone Modifications in Lanthipeptides
Ayikpoe, R.S.; van der Donk, W.A.* (2021) in Methods in Enzymology; Petersson, E.J., Ed.; Academic Press: Cambridge, 573-621.
308. Structural Analysis of Class I Lanthipeptides from Pedobacter lusitanus NL19 Reveals Unusual Ring Topology
Bothwell, I.R.: Caetano, T.; Sarksian, R.; Mendo, S.*; van der Donk, W.A.* (2021) ACS Chem. Biol., 16, 1019-1029.
307. Biosynthesis of Fosfomycin in Pseudomonads Reveals a New Enzymatic Activity in the Metallohydrolase Superfamily
Simon, M.A.; Ongpipattanakul, C.; Nair, S.K.*: van der Donk, W.A.* (2021) Proc. Natl. Acad. Sci. USA, 118, e2019863118.
306. Overall Retention of Methyl Stereochemistry During B12-Dependent Radical SAM Methyl Transfer in Fosfomycin Biosynthesis
McLaughlin, M.I.; Pallitsch, K.; Wallner, G.; van der Donk, W.A.*; Hammerschmidt, F.* (2021) Biochemistry, 60, 1587-1596.
305. Engineering of New-to-Nature Ribosomally Synthesized and Post-Translationally Modified Peptide Natural Products
Wu, C.; van der Donk, W.A.* (2021) Curr. Opin. Biotechnol., 69, 221-231.
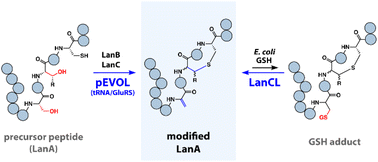
304. LanCLs Add Glutathione to Dehydroamino Acids Generated at Phosphorylated Sites in the Proteome
Lai, K.-Y.; Galan, S.R.G.; Zeng, Y.; He, C.; Riedl, J.; Raj, R.; Chooi, K.P.; Garg, N.; Jones, L.H.; Hutchings, G.J.; Mohammed, S.; Nair, S.K.; Chen, J.*; Davis, B.G.*; van der Donk, W.A.* (2021) Cell, 184, 2680-2695.
303. MicroED in Natural Product and Small Molecule Research
Danelius, E.; Halaby, S.; van der Donk, W.A.*; Gonen, T.* (2021) Nat. Prod. Rep., 38, 423-431.
302. Mechanisms and Evolution of Diversity-Generating RiPP Biosynthesis
Le, T.; van der Donk, W.A.* (2021) Trends Chem., 3, 266-278.
301. New Developments in RiPP Discovery, Enzymology and Engineering
Montalbán-López, M.; Scott, T.A.; Ramesh, S.; Rahman, I.R.; van Heel, A.; Viel, J.H.; Bandarian, V.; Elke Dittmann, E.; Genilloud, O.; Goto, Y.; Grande Burgos, M.J.; Hill, C.; Kim, S.; Koehnke, J.; Latham, J.A.; Link A.J.; Martínez, B.; Nair, S.K.; Nicolet, Y.; Rebuffat, S.; Sahl, H.-G.; Sareen, D.; Schmidt, E.W.; Schmitt, L.; Severinov, K.; S Süssmuth, R.D.; Truman, A.W.; Wang, H.; Weng, J.-K.; van Wezel, G.P.; Zhang, Q.; Zhong, J.; Piel, J.;* Mitchell, D.A;* Kuipers, O.P.;* van der Donk, W.A.* (2021) Nat. Prod. Rep., 38, 130-239.
2020
300. Structural Determinants of Macrocyclization in Substrate-Controlled Lanthipeptide Biosynthetic Pathways
Bobeica, S.C.; Zhu, L.; Acedo, J.Z.; Tang, W.; van der Donk, W.A.* (2020) Chem. Sci., 11, 12854-12870.
299. Discovery and Characterization of a Class IV Lanthipeptide with a Non-Overlapping Ring Pattern
Ren, H.; Shi, C.; Bothwell, I.R.; van der Donk, W.A.*; Zhao, H.* (2020) ACS Chem. Biol., 15, 1642-1649.
298. Recent Progress in Lanthipeptide Biosynthesis, Discovery, and Engineering
An, L.; van der Donk, W.A.*(2019) Comprehensive Natural Products III: Chemistry and Biology; Liu, H.-W., Begley, T.P., Eds.; Elsevier: Oxford, 2019; pp 119-165.
297. Substrate Recognition by the Class II Lanthipeptide Synthetase HalM2
Rahman, I.R.; Acedo, J.Z; Liu, X.R.; Zhu, L.; Arrington, J.; Gross, M.L.; van der Donk, W.A.* (2020) ACS Chem. Biol., 15, 1473-1486.
296. Precursor Peptide-Targeted Mining of More Than One Hundred Thousand Genomes Expands the Lanthipeptide Natural Product Family
Walker, M.*; Eslami, S.M.; Hetrick, K.J.; Ackenhusen, S.E.; Mitchell, D.A.; van der Donk, W.A. (2020) BMC Genom., 21, 387.
295. Bacteroidetes Are a Rich Source of Novel Lanthipeptides: The Case Study of Pedobacter lusitanus
Caetano, T.*; van der Donk, W.A.; Mendo, S. (2020) Microbiol. Res., 235, 126441.
294. The Fellowship of the Rings: Macrocyclic Antibiotic Peptides Reveal Anti-Gram Negative Target
McLaughlin, M.; van der Donk, W.A.* (2020) Biochemistry, 59, 343-345.
293. Characterization of a dehydratase and methyltransferase in the biosynthesis of a ribosomally-synthesized and post-translationally modified peptide in Lachnospiraceae
Huo, L.; Zhao, X.; Acedo, J.Z.; Estrada, P.; Nair, S.K.; van der Donk, W.A.* (2020) ChemBioChem, 21, 190-199.
2019
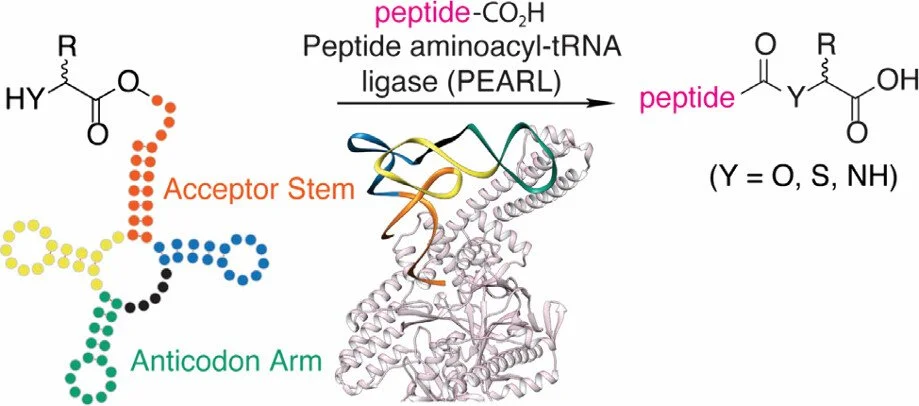
292. Nonribosomal Peptide Extension by a Peptide Amino-acyl tRNA Ligase
Zhang, Z.; van der Donk, W.A.* (2019) J. Am. Chem. Soc., 141, 19625-19633.
291. Bacteriophage Targeting of Gut Bacterium Attenuates Alcoholic Liver Disease
Duan, Y.; Llorente, C.; Brand, K.; Chu, H.; Jiang, L.; Lang, S.; Torralba, M.; Shao, Y.; Liu, J.; Hernandez-Morales, A.; Lessor, L.; Rahman, I.R.; Miyamoto, Y.; Ly, M.; Sun, W.; Kiesel, R.; Hutmacher, F.; Lee, S.; Ventura-Cots, M.; Bosques-Padilla, F.; Verna, E.C.; Abraldes, J.G.; Brown Jr, R.S.; Vargas, V.; Altamirano, J.; Caballería, J.; Shawcross, D.; Ho, S.B.; Louvet, A.; Lucey, M.R.; Mathurin, P.; Garcia-Tsao, G.; Bataller, R.; Tu, X.M.; Eckmann, L.; van der Donk, W.A.; Young, R.; Trevor, L.; Pride, D.; Stärkel, P.; Fouts, D.E.; Schnabl, B.* (2019) Nature, 575, 505-511.
290. O-Methyltransferase-mediated Incorporation of a β-Amino Acid in Lanthipeptides
Acedo, J.; Bothwell, I.; An, L.; Trouth, A.; Frazier, C.; van der Donk, W.A.* (2019) J. Am. Chem. Soc., 141, 16790-16801.
289. Temperature-independent kinetic isotope effects as evidence for a Marcus-like model of hydride tunneling in phosphite dehydrogenase
Howe, G.W.*; van der Donk, W.A.* (2019) Biochemistry, 58, 4260-4268.
288. Characterization of Glutamyl-tRNA Dependent Dehydratases using Non-Reactive Substrate Mimics
Bothwell, I.R.; Cogan, D.P.; Kim, T.; Reinhardt, C.J.; van der Donk, W.A.*; Nair, S.K.* (2019) Proc. Natl. Acad. Sci. USA, 35, 17245-17250.
287. Use of a Scaffold Peptide in the Biosynthesis of Amino Acid Derived Natural Products
Ting, C.P.; Funk, M.A.; Halaby, S.L.; Zhang, Z.; Gonen, T.; van der Donk, W.A.* (2019) Science, 365, 280-284.
286. Mechanistic Studies of the Kinase Domains of Class IV Lanthipeptide Synthetases
Hegemann, J.D.; Shi, L.; Gross, M.L.*: van der Donk, W.A.* (2019) ACS Chem. Biol., 14, 1583-1592.
285. Assessing the Flexibility of the Prochlorosin 2.8 Scaffold for Bioengineering Applications
Hegemann, J.D.; Bobeica, S.C.; Walker, M.C.; Bothwell, I.R.; van der Donk, W.A.* (2019) ACS Synth. Biol., 8, 1204-1214.
284. Insights into AMS/PCAT Transporters from Biochemical and Structural Characterization of a Double Glycine Motif Protease
Bobeica, S.C.; Dong, S.-H.; Huo, L.; Mazo, N.; McLaughlin, M.I.; Jiménez-Osés, G.; Nair, S.K.*; van der Donk, W.A.* (2019) eLife, 8, e42305.
283. Investigations into the mechanism of action of sublancin
Wu, C.; Biswas, S.; Garcia De Gonzalo, C.V.; van der Donk, W.A* (2019) ACS Infect. Dis., 5, 454-459.
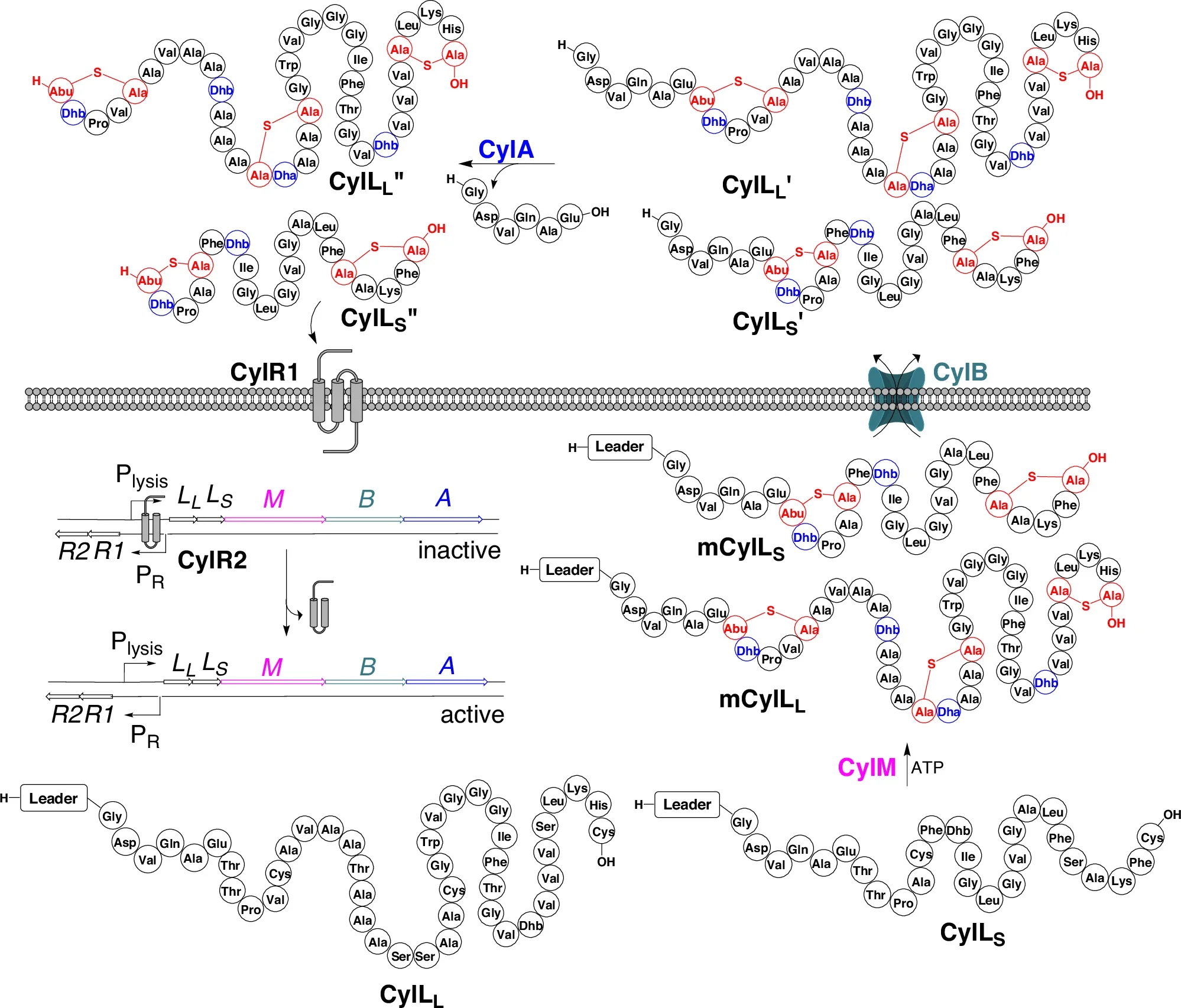
282. CylA is a Sequence-Specific Protease Involved in Toxin Biosynthesis
Tang, W.; Bobeica, S.C.; Wang, L.; van der Donk* (2019) J. Ind. Microbiol. Biotechnol., 46, 537-549.
281. Use of the Dehydrophos Biosynthetic Enzymes to Prepare Antimicrobial Analogs of Alaphosphin
Bougioukou, D.J.; Ting, C.P.; Peck, S.C.; Mukherjee, S.; van der Donk, W.A.* (2019) Org. Biomol. Chem., 17, 822-829.
2018
280. 18O Kinetic Isotope Effects Reveal an Associative Transition State for Phosphite Dehydrogenase Catalyzed Phosphoryl Transfer
Howe, G.W.; van der Donk, W.A.* (2018) J. Am. Chem. Soc., 140, 17820-17824.
279. Rapid Discovery of Glycocins through Pathway Refactoring in Escherichia coli
Ren H.; Biswas, S.; Ho, S.; van der Donk, W.A.*; Zhao, H.* (2018) ACS Chem. Biol., 13, 2966-2972.
278. Rapid structure-activity screening of lanthipeptide analogs via in-colony removal of leader peptides in Escherichia coli
Si, T.; Tian, Q.; Min, Y.; Zhang, L.; Sweedler, J.; van der Donk, W.A.*; Zhao, H.* (2018) J. Am. Chem. Soc., 140, 11884-11888.
277. Glutamic acid is a carrier for hydrazine during the biosyntheses of fosfazinomycin and kanamycin
Wang, K.K.A.; Ng, T.L.; Wang, P.; Huang, Z.; Balskus, E.P.*, van der Donk, W.A.* (2018) Nat. Comm., 9, 3687.
276. Substrate-assisted Enzymatic Formation of Lysinoalanine in Duramycin
An, L.; Cogan, D.P.; Navo, C.D.; Jiménez-Osés, G.; Nair, S.K.*; van der Donk, W.A.* (2018) Nat. Chem. Biol., 14, 28-933.
275. Lanthionine Synthetase C-like Protein 2 (LanCL2) is Important for Adipogenic Differentiation
Dutta, D.; Lai, K.-Y.; Reyes-Ordoñez, A.; Chen, J.; van der Donk, W.A* (2018) J. Lipid Res., 8, 1433-1445.
274. Stereospecific Radical-mediated B12-dependent Methyl Transfer by the Fosfomycin Biosynthesis Enzyme Fom3
McLaughlin, M.I.; van der Donk, W.A.* (2018) Biochemistry, 57, 4967-4971.
273. Elucidation of the Roles of Conserved Residues in the Biosynthesis of the Lasso Peptide Paeninodin
Hegemann, J.D.; Schwalen, C.J.; Mitchell, D.A.*; van der Donk, W.A.* (2018) Chem. Comm., 54, 9007-9010.
272. The Enzymology of Prochlorosin Biosynthesis
Bobeica, S.C.; van der Donk, W.A.* (2018) Methods Enzymol., 604, 165-203.
271. Investigation of Substrate Recognition and Biosynthesis in Class IV Lanthipeptide Systems
Hegemann, J.D.; van der Donk, W.A.* (2018) J. Am. Chem. Soc., 140, 5743-5754.
270. Incorporation of Nonproteinogenic Amino Acids in Class I and II Lantibiotics
Kakkar, N.; Perez, J.; Liu, W.R.; Jewett, M.; van der Donk, W.A.* (2018) ACS Chem. Biol., 20, 951-957.
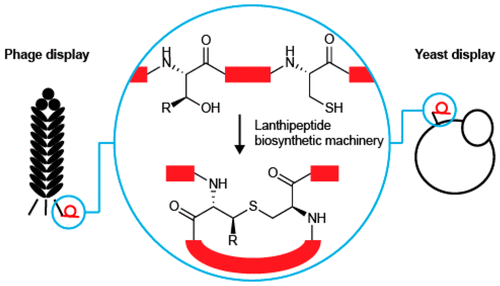
269. Development and Application of Yeast and Phage Display of Diverse Lanthipeptides
Hetrick, K.J.; Walker, M.C; van der Donk, W.A.* (2018) ACS Cent. Sci., 4, 458–467.
268. Cyclic Peptides: from Bioorganic Synthesis to Applications
Koehnke, J.; Naismith, J.; van der Donk, W. A. (2018) Royal Society of Chemistry: Cambridge, UK.
267. Investigation of Amide Bond Formation during Dehydrophos Biosynthesis
Ulrich, E.C.; Bougioukou, D.J.; van der Donk, W.A.* (2018) ACS Chem. Biol., 13, 537-541.
266. A lanthipeptide library used to identify inhibitors of a protein-protein interaction
Yang, X.; Lennard, K.R.: He, C.; Walker, M.C.; Ball, A.T.; Doigneaux, C.; Tavassoli, A.*; van der Donk, W.A.* (2018) Nat. Chem. Biol., 14, 375-380.
265. Characterization of Leader Peptide Binding during Catalysis by the Nisin Dehydratase NisB
Repka, L.M.; Hetrick, K.J.; Chee, S.H.; van der Donk, W.A.* (2018) J. Am. Chem. Soc., 140, 4200-4203.